Efficient querying of genomic databases.
Project Links
Meta
Author: Laura Luebbert
Maintainer: Laura Luebbert
Requires Python: >=3.7
Classifiers
Environment
- Console
Framework
- Jupyter
Intended Audience
- Science/Research
License
- OSI Approved :: BSD License
Operating System
- OS Independent
Programming Language
- Python :: 3.7
- Python :: 3.8
- Python :: 3.9
- Python :: 3.10
- Python :: 3.11
- Python :: 3.12
Topic
- Scientific/Engineering :: Bio-Informatics
- Utilities
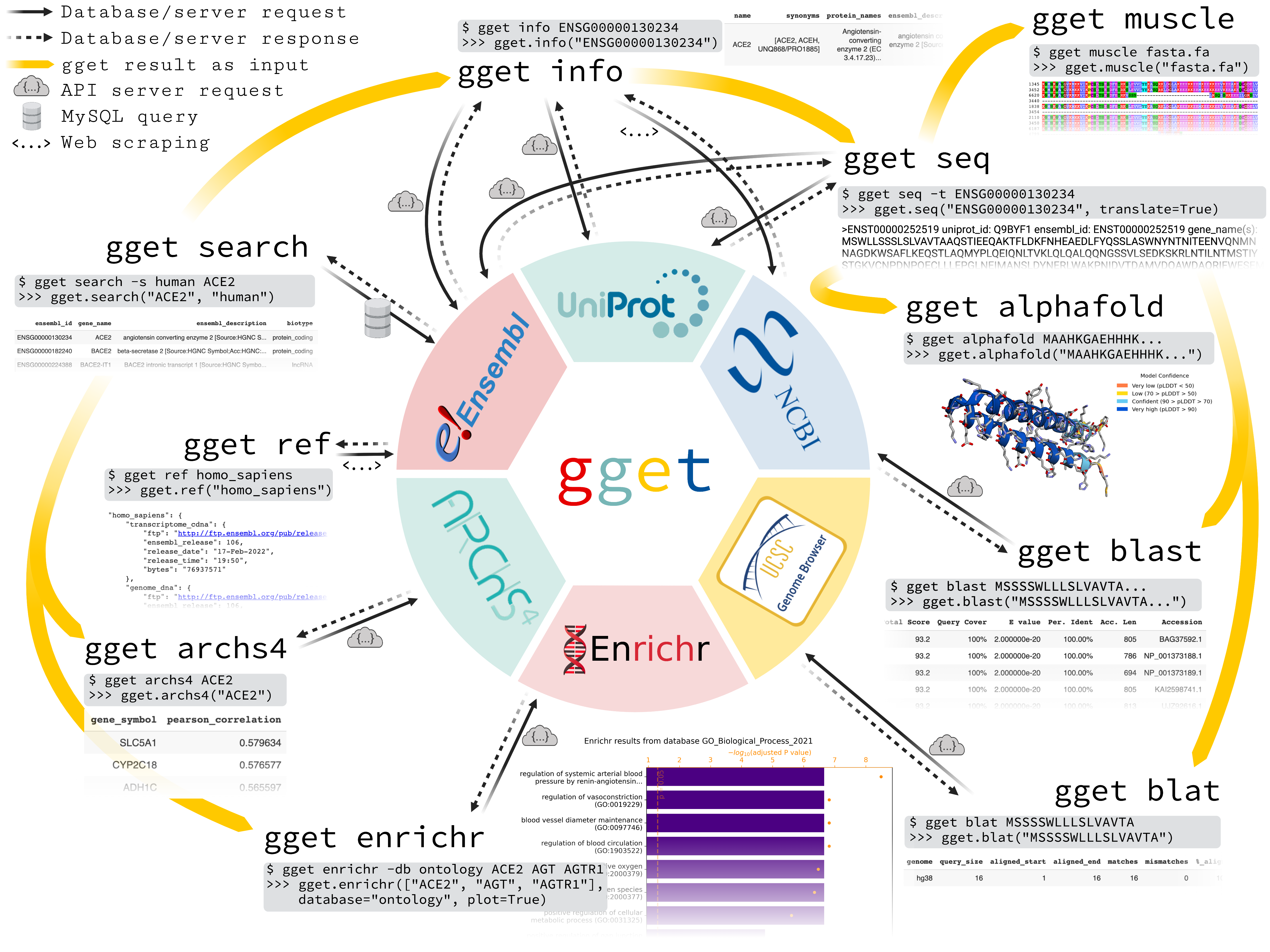
gget
gget is a free, open-source command-line tool and Python package that enables efficient querying of genomic databases. gget consists of a collection of separate but interoperable modules, each designed to facilitate one type of database querying in a single line of code.
If you use gget in a publication, please cite*:
Luebbert, L., & Pachter, L. (2023). Efficient querying of genomic reference databases with gget. Bioinformatics. https://doi.org/10.1093/bioinformatics/btac836
Read the article here: https://doi.org/10.1093/bioinformatics/btac836
Installation
uv pip install gget
or
pip install --upgrade gget
For use in Jupyter Lab / Google Colab:
# Python
import gget
🔗 Manual
🪄 Quick start guide
Command line:
# Fetch all Homo sapiens reference and annotation FTPs from the latest Ensembl release
$ gget ref homo_sapiens
# Get Ensembl IDs of human genes with "ace2" or "angiotensin converting enzyme 2" in their name/description
$ gget search -s homo_sapiens 'ace2' 'angiotensin converting enzyme 2'
# Look up gene ENSG00000130234 (ACE2) and its transcript ENST00000252519
$ gget info ENSG00000130234 ENST00000252519
# Fetch the amino acid sequence of the canonical transcript of gene ENSG00000130234
$ gget seq --translate ENSG00000130234
# Quickly find the genomic location of (the start of) that amino acid sequence
$ gget blat MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# BLAST (the start of) that amino acid sequence
$ gget blast MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# Align multiple nucleotide or amino acid sequences against each other (also accepts path to FASTA file)
$ gget muscle MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
# Align one or more amino acid sequences against a reference (containing one or more sequences) (local BLAST) (also accepts paths to FASTA files)
$ gget diamond MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS -ref MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS
# Use Enrichr for an ontology analysis of a list of genes
$ gget enrichr -db ontology ACE2 AGT AGTR1 ACE AGTRAP AGTR2 ACE3P
# Get the human tissue expression of gene ACE2
$ gget archs4 -w tissue ACE2
# Get the protein structure (in PDB format) of ACE2 as stored in the Protein Data Bank (PDB ID returned by gget info)
$ gget pdb 1R42 -o 1R42.pdb
# Find Eukaryotic Linear Motifs (ELMs) in a protein sequence
$ gget setup elm # setup only needs to be run once
$ gget elm -o results MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS
# Fetch a scRNAseq count matrix (AnnData format) based on specified gene(s), tissue(s), and cell type(s) (default species: human)
$ gget setup cellxgene # setup only needs to be run once
$ gget cellxgene --gene ACE2 SLC5A1 --tissue lung --cell_type 'mucus secreting cell' -o example_adata.h5ad
# Predict the protein structure of GFP from its amino acid sequence
$ gget setup alphafold # setup only needs to be run once
$ gget alphafold MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK
Python (Jupyter Lab / Google Colab):
import gget
gget.ref("homo_sapiens")
gget.search(["ace2", "angiotensin converting enzyme 2"], "homo_sapiens")
gget.info(["ENSG00000130234", "ENST00000252519"])
gget.seq("ENSG00000130234", translate=True)
gget.blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.muscle(["MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"])
gget.diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS")
gget.enrichr(["ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"], database="ontology", plot=True)
gget.archs4("ACE2", which="tissue")
gget.pdb("1R42", save=True)
gget.setup("elm") # setup only needs to be run once
ortho_df, regex_df = gget.elm("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget.setup("cellxgene") # setup only needs to be run once
gget.cellxgene(gene = ["ACE2", "SLC5A1"], tissue = "lung", cell_type = "mucus secreting cell")
gget.setup("alphafold") # setup only needs to be run once
gget.alphafold("MSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTFSYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYK")
Call gget from R using reticulate:
system("pip install gget")
install.packages("reticulate")
library(reticulate)
gget <- import("gget")
gget$ref("homo_sapiens")
gget$search(list("ace2", "angiotensin converting enzyme 2"), "homo_sapiens")
gget$info(list("ENSG00000130234", "ENST00000252519"))
gget$seq("ENSG00000130234", translate=TRUE)
gget$blat("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget$blast("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS")
gget$muscle(list("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", "MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS"), out="out.afa")
gget$diamond("MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLAS", reference="MSSSSWLLLSLVEVTAAQSTIEQQAKTFLDKFHEAEDLFYQSLLAS")
gget$enrichr(list("ACE2", "AGT", "AGTR1", "ACE", "AGTRAP", "AGTR2", "ACE3P"), database="ontology")
gget$archs4("ACE2", which="tissue")
gget$pdb("1R42", save=TRUE)
More tutorials
0.29.3
Sep 11, 2025
0.29.2
Jul 03, 2025
0.29.1
Apr 21, 2025
0.29.0
Sep 26, 2024
0.28.6
Jun 03, 2024
0.28.5
May 30, 2024
0.28.4
Feb 01, 2024
0.28.3
Jan 22, 2024
0.28.2
Nov 16, 2023
0.28.0
Nov 12, 2023
0.27.9
Aug 07, 2023
0.27.8
Jul 12, 2023
0.27.7
May 16, 2023
0.27.6
May 02, 2023
0.27.5
Apr 06, 2023
0.27.4
Mar 19, 2023
0.27.3
Mar 11, 2023
0.27.2
Jan 01, 2023
0.27.1
Dec 30, 2022
0.27.0
Dec 10, 2022
0.3.13
Nov 11, 2022
0.3.12
Nov 10, 2022
0.3.11
Sep 07, 2022
0.3.10
Sep 02, 2022
0.3.9
Aug 25, 2022
0.3.8
Aug 12, 2022
0.3.7
Aug 09, 2022
0.3.5
Aug 06, 2022
0.3.4
Aug 06, 2022
0.3.3
Aug 05, 2022
0.3.1
Aug 05, 2022
0.3.0
Aug 04, 2022
0.2.7
Jul 29, 2022
0.2.6
Jul 08, 2022
0.2.5
Jun 30, 2022
0.2.4
Jun 29, 2022
0.2.3
Jun 27, 2022
0.2.2
Jun 24, 2022
0.2.1
Jun 09, 2022
0.2.0
Jun 08, 2022
0.1.2
Jun 03, 2022
0.1.1
May 28, 2022
0.1.0
May 25, 2022
0.0.24
May 17, 2022
0.0.23
May 17, 2022
0.0.22
May 10, 2022
0.0.17
Mar 02, 2022
0.0.16
Mar 02, 2022
0.0.6
Feb 26, 2022
0.0.5
Feb 25, 2022
0.0.4
Feb 22, 2022
Wheel compatibility matrix
Files in release
Extras:
None
Dependencies:
numpy
(>=1.17.2)
pandas
(>=1.0.0)
requests
(>=2.22.0)
ipython
matplotlib
mysql-connector-python
(>=8.0.32)
beautifulsoup4
(>=4.10.0)
ipywidgets
tqdm
lxml





